N-terminal Amino Acid Sequencing Analysis by MALDI-TOF MS/Protein Sequencer
Mass spectrometry has become an indispensable tool for researchers looking to sequence peptides. Although effective in many cases, sequencing by In Source Decay (ISD) faces a few challenges its ability to provide reliable sequence information including isobaric amino acids, database dependency and low molecular weight interferences.
Traditional Edman sequencing avoids mass dependency and the use of databases by analyzing each amino acid from the N-terminus one at a time in sequence. Unfortunately, Edman has its own limitations in providing high sequence coverage.
This technical note investigates the benefits of combining the intact mass and sequencing information from the MALDI-8020 (ISD) with the N- terminal sequence obtained from the PPSQ-50A gradient system (Edman). Using the combined information allows investigators the ability to obtain a more complete picture of their proteins and peptides of interest.
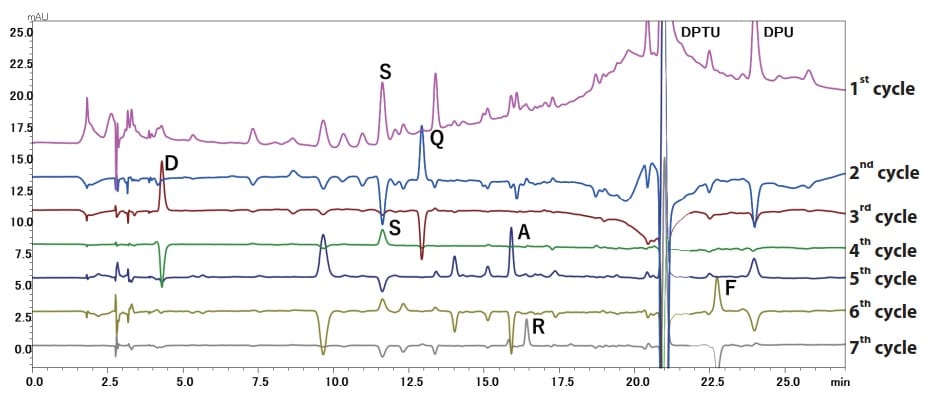
Fig. 1 Subtracted Chromatograms from First Seven Cycles Using the PPSQ-50A Gradient System
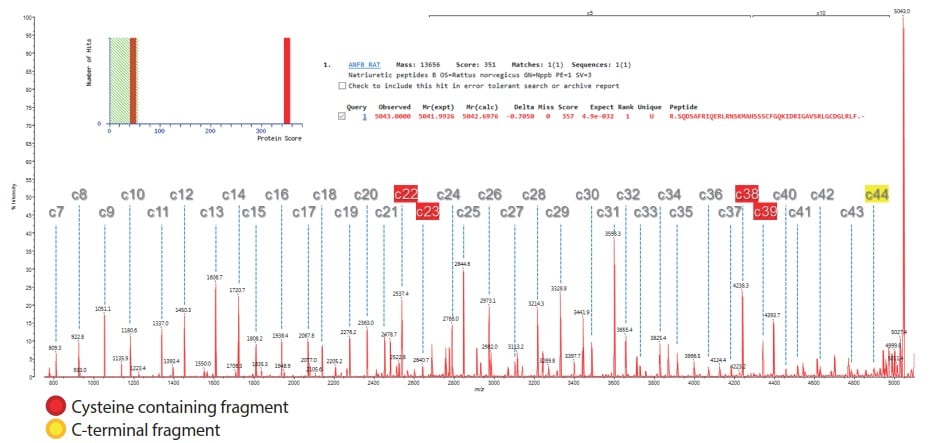
Fig. 2 ISD Spectrum of BNP Using MALDI-8020 with DAN Matrix. MASCOT Identification Shown Inset
■ Related Application News ■
Protein Sequencer

Features:
The PPSQ-51A/53A continues the tradition of providing reliable and sensitive N-terminal protein sequencing to researchers through automated Edman Degradation. Whether transitioning over from another sequencer or adopting the technique for the first time, the intuitive software and robust Shimadzu hardware will make protein sequencing easy on your laboratory.



