MALDImini-1 - Features
MALDI Digital Ion Trap Mass Spectrometer
"Minimum" footprint
The MALDI-MS that can be installed anywhere

Until now, devices for MSn have had strenuous installation requirements, but the MALDImini-1’s compact size and power requirements make it easy to install almost anywhere. Easy access to the instrument allows users to quickly acquire data and check their results, creating a more convenient workflow right on the bench.
"Minimum" time
Begin taking measurements quickly without fuss
MALDI lends itself to quick analysis through simple sample preparation and acquisition. Once the sample is loaded, users can acquire a spectrum in a few minutes at the click of a button.
With such a smooth and quick setup, it is possible to carry out detailed structural analyses in a very short time span.
"Minimum" sample volume
MSn analysis is possible over a wide mass range, even with microquantity samples
By combining a MALDI ion source with Digital Ion Trap (DIT) technology, it is possible to carry out high-sensitivity MS and MSn analysis even on micro-quantity samples. Not only can the mass of various molecules be checked, but it is also possible to carry out a wide range of analyses such as identifying proteins and investigating the structure of glycans and glycopeptides.
■Identifying protein digests
By carrying out MS and MS/MS measurements on protein digests, it is possible to determine the identify of a protein.
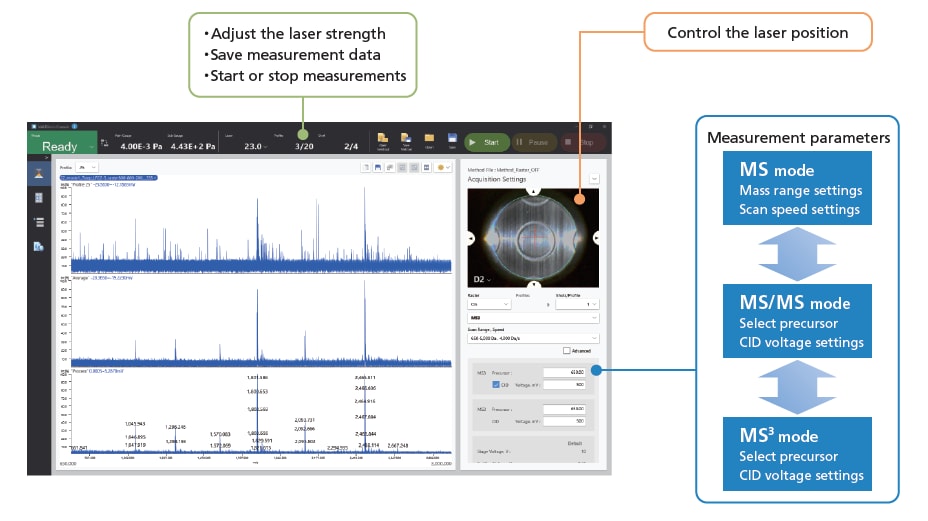
■MALDImini Console
With the dedicated control software for MALDImini-1, you can take measurements just by setting simple analysis parameters. Switch quickly and easily between MS, MS/MS and MS3 modes for seamless analysis. Data can be exported in mzML or mzXML formats.
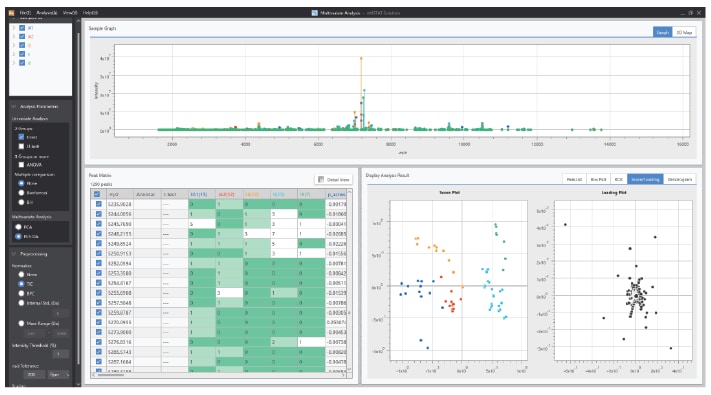
■Compatibility with Software Solutions
Powerful processing tools can be used to analyze your exported mzML or mzXML data, such as MASCOT for identifying protein digests, SimGlycan™ for processing MSn data for glycans, and eMSTAT Solution™ for statistical analysis.
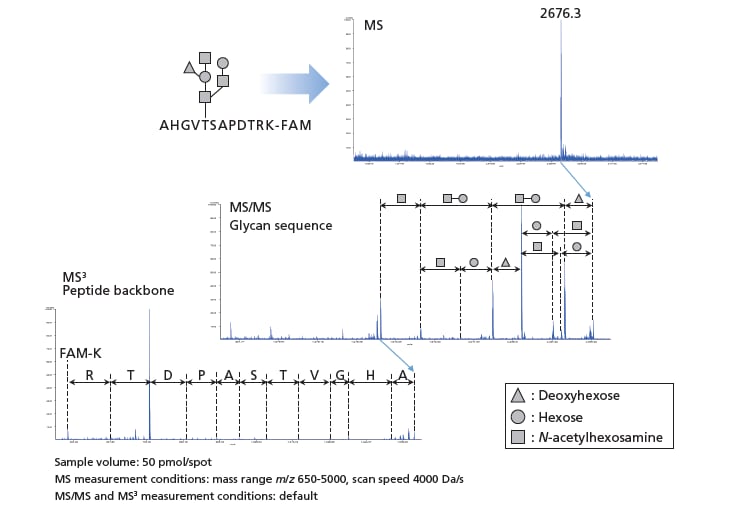
■Investigating glycopeptide structure
Through MS, MS/MS and MS3 measurements of synthetic O-linked glycopeptides, the glycan sequence can be found from MS/MS data and the amino acid sequence of the peptide backbone can be found from MS3 data.
MALDImini, eMSTAT Solution and FlexiMass are trademarks of Shimadzu Corporation.
MASCOT is a registered trademark of Matrix Science Ltd.
SimGlycan is a trademark of PREMIER Biosoft International.