LC/MS/MS MRM Library for Metabolic Enzymes in Yeast
For LabSolutions™ LCMS Software
Includes MRM Analysis Conditions Necessary for Analyzing Metabolic Enzymes

An enormous number of MRM conditions need to be evaluated in order to analyze a large number of proteins.
This MRM library includes such analysis conditions, greatly facilitating the initial preparation for analysis. By allowing the use of proven methods, it greatly reduces the time needed for method development.
Ideal for the Analysis of Enzymes Related to Primary Metabolism Derived from Yeast
This product provides a library consisting of 3,584 MRM transitions, including stable isotopes. It covers all 498 trypsin digested peptides of 228 types of enzymes derived from budding yeast, which is used for the production of bioethanol or other materials, or as a model organism for basic research. This library enables a variety of enzyme measurements, including those related to the major metabolic pathways of glycolysis, the TCA cycle, the pentose phosphate cycle, and amino acid metabolism.
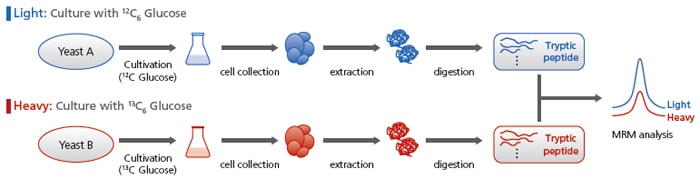
Comparative Analyses Using Stable Isotopes
The MRM analysis conditions for 13C-labeled peptides, for all MRM transitions, are listed. Using these conditions enables comparative analyses.
For example, a gene deficient yeast group grown with unlabeled glucose can be compared with a control yeast group grown with labeled glucose.
Easy-to-Customize Methods
The method parameter list included in this package can be used to create methods that analyze targeted components only.
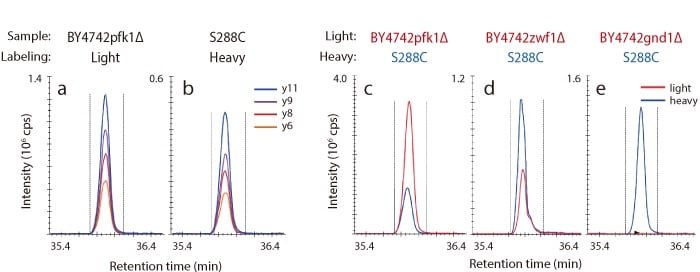
MRM Analysis of Gnd1p Trypsin Digested Peptides in Gene-Disrupted Strains
Shown below are representative chromatograms for a BY4742pfk1Δ strain (light) grown with unlabeled glucose (a), and a S288C strain (heavy) grown with 13C-labeled glucose (b). Additionally, TIC chromatograms of Gnd1p in gene-disrupted strains are shown in (c), (d), and (e). In GND1 disrupted strains, Gnd1p was not detected, whereas in PFK1 disrupted strains, large numbers of Gnd1p were detected.
|
Reference) Matsuda F, Ogura T, Tomita A, Hirano I, Shimizu H. Nano-scale liquid chromatography coupled to tandem mass spectrometry using the multiple reaction monitoring mode based quantitative platform for analyzing multiple enzymes associated with central metabolic pathways ofSaccharomyces cerevisiaeusing ultra fast mass spectrometry.J Biosci Bioeng. 2015 Jan;119(1):117-20. |
Registered List of Metabolic Enzymes
AAT1
AAT2
ACH1
ACO1
ACO2
ACS1
ACS2
ADE1
ADE12
ADE13
ADE16
ADE17
ADE2
ADE4
ADE5,7
ADE6
ADH1
ADH2
ADH3
ADH4
ADH6
ADK1
AGX1
ALD3
ALD4
ALD5
ALD6
ALT1
ALT2
ARG1
ARG2
ARG3
ARG4
ARG5,6
ARG8
ARO1
ARO2
ARO3
ARO4
ARO7
ARO8
ASN1
ASN2
ASP1
ATH1
BAT1
BAT2
BNA3
BNA5
CAR1
CAR2
CDC19
CIT1
CIT2
CIT3
CPA1
CPA2
CYS3
CYS4
DAK1
DAL7
DUR1,2
ECM17
ECM40
ENO1
ENO2
ERG10
ERG13
ERG20
ERR
FBA1
FBP1
FRD1
FUM1
GAD1
GAL1
GAL10
GAL7
GCV1
GCV2
GCY1
GDB1
GDH1
GDH2
GDH3
GLC3
GLK1
GLN1
GLT1
GLY1
GND1
GND2
GPD1
GPD2
GPH1
GPM1
GPM2
GPM3
GSY1
GSY2
GUA1
GUK1
GUT2
HIS1
HIS3
HIS4
HIS5
HIS6
HIS7
HOM2
HOM3
HOM6
HOR2
HXK1
HXK2
ICL1
IDH1
IDH2
IDP1
IDP2
IDP3
ILV1
ILV2
ILV3
ILV5
IMD2
IMD4
KGD1
KGD2
LAT1
LEU1
LEU2
LEU4
LPD1
LSC1
LSC2
LYS1
LYS12
LYS2
LYS20
LYS21
LYS4
LYS9
MAE1
MDH1
MDH2
MDH3
MET10
MET14
MET16
MET17
MET2
MET22
MET3
MET6
MHT1
MLS1
MVD1
NQM1
NTH1
PCK1
PDA1
PDB1
PDC1
PDC5
PDC6
PDE1
PFK1
PFK2
PGI1
PGK1
PGM1
PGM2
PRO1
PRO2
PRO3
PRS1
PRS2
PRS3
PRS4
PRS5
PYC1
PYC2
RHR2
RKI1
RNR2
RNR4
RPE1
SAH1
SAM1
SAM2
SAM4
SDH1
SDH2
SDH3
SDH4
SER1
SER2
SER3
SER33
SFA1
SHM2
SOL3
SOL4
TAL1
TDH1
TDH2
TDH3
THR1
THR4
TKL1
TKL2
TPI1
TPS1
TPS2
TPS3
TRP1
TRP2
TRP3
TRP5
TSL1
UGA1
UGA2
UGP1
URA2
YNK1
YPR1
ZWF1


