This Is The Power Of AI - Discover LabSolutions MD Now
MALDI-MS Protein Profiling of Chemoresistance in Extracellular Vesicles of Cancer Cells
Introduction

As part of a collaboration with the Medical University of Vienna, this project aimed to make the detection of chemoresistance biomarkers faster and less invasive for patients undergoing chemotherapy. Faster detection of chemoresistance will improve cancer therapy and therefore improve cancer survival rates.
Cancer cells communicate with the whole organism via extracellular vesicles (EVs), which circulate the body and propagate molecular information in support of the malignant phenotype.
Colon cancer lymph node metastasis were cultured with progressive levels of fluorouracil to build increasing levels of chemoresistance. EVs were harvested from cell culture supernatant by ultracentrifugation (see Fig. 1) to serve as a model for circulating cancer cell-derived biomarker carriers from body fluids (i.e., liquid biopsy).
In this work, differential expression of proteins in EVs, measured by MALDI-TOF-MS, as the result of an increasing chemoresistance of their parent cells was observed (1).
Read Journal Article: MALDI-MS Protein Profiling of Chemoresistance in Extracellular Vesicles of Cancer Cells (2018)
Samples and Methods
EVs and cell extracts derived from primary colon tumour, lymph node metastasis subclones resistant to 5, 25 and 125 µM 5-fluorouracil (FU) were provided by Dr. Gerald Stübiger at the Medical University of Vienna. Using the MALDI-8020 MALDI-TOF-MS system, protein mass spectra were recorded using CHCA in the range m/z 2,000−25,000 and subjected to multivariate data analysis (eMSTAT Solution™ software, Shimadzu), see Fig. 1. By using hierarchical clustering, PLS-DA, discriminatory protein patterns of the EVs from different cell populations were obtained.

Results
A representative mass spectrum for one of the EV samples is shown in Fig. 2. The sensitivity of the MALDIMS based assay was in the low μg/mL (≈1.2−5 × 1010 particles/mL) range (data not shown).
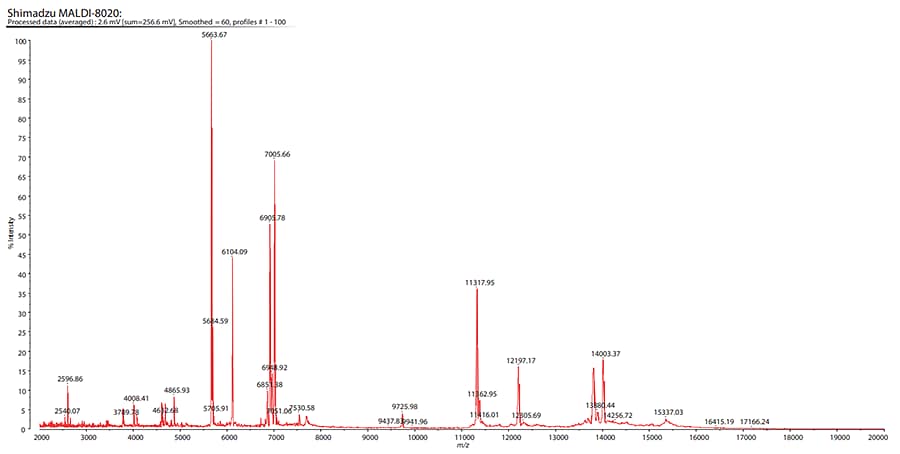
Fig. 2 Spectrum of Extracellular Vesicle Protein Expression Derived from the Primary Colon Cancer
Following data acquisition, the peaklists were imported into eMSTAT Solution for analysis. Replicates for each sample type were grouped together. The software makes it easy for the user to identify the differentiating peaks. The score plot (Fig. 3) shows the distinction between the naive and the fluorouracilresistant sample groups. The fluorouracil-resistant groups share sufficient similarities to be grouped together whilst exhibiting sufficient distinction for those groups to be separated.
Peaks in the range m/z 2,000-7,000 were found to be the most helpful in characterising chemoresistance.
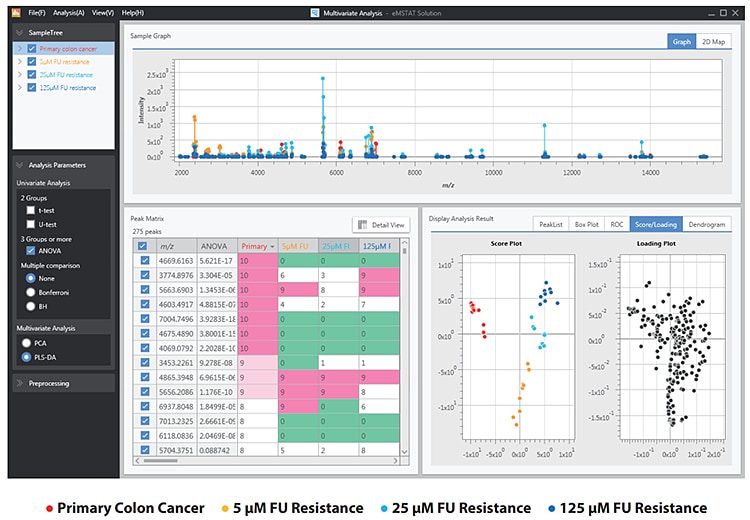
Fig. 3 PLS-DA Analysis of the 4 Vesicle Expression Groups
Conclusion
This work highlights the utility of the MALDI-8020 combined with eMSTAT Solution software to facilitate biomarker discovery.
The MALDI-MS protein profiling approach shows the potential to serve as novel tool for minimally invasive cancer diagnostics and chemotherapy monitoring in the future, e.g., for early detection of therapy resistance without biopsy.


