Protein Sequence Analysis by In-Source Decay Using a Benchtop MALDI-TOF Mass Spectrometer
MALDI-TOF mass spectrometers are frequently used for molecular weight measurement and identification of proteins. To identify proteins using a mass spectrometer, generally tryptic digestion is required for preparation; however, if the protein is isolated and purified, by detecting the ions produced by fragmentation within an ion source (ISD, In-Source Decay), sequence analysis of intact proteins can be performed with no need to digest the protein with trypsin.
When measuring molecular weights of proteins using MALDI, reagents (matrices) comprising sinapinic acid are normally used; however, when performing sequence analysis by ISD, 1,5-diaminonaphthalene (DAN) is used as a matrix. By conducting a database search using the fragment ions produced by ISD, e.g. using Mascot, proteins can be identified. In addition, by conducting a homology search for amino acid sequence information obtained from the masses of fragment ions, it is possible to infer the proteins which were not matched against the protein database.
This article introduces an example of measuring molecular weights of proteins and performing sequence analysis of proteins by ISD using a benchtop MALDI-TOF mass spectrometer "MALDI-8020".
Measurement Example of Protein Molecular Weight
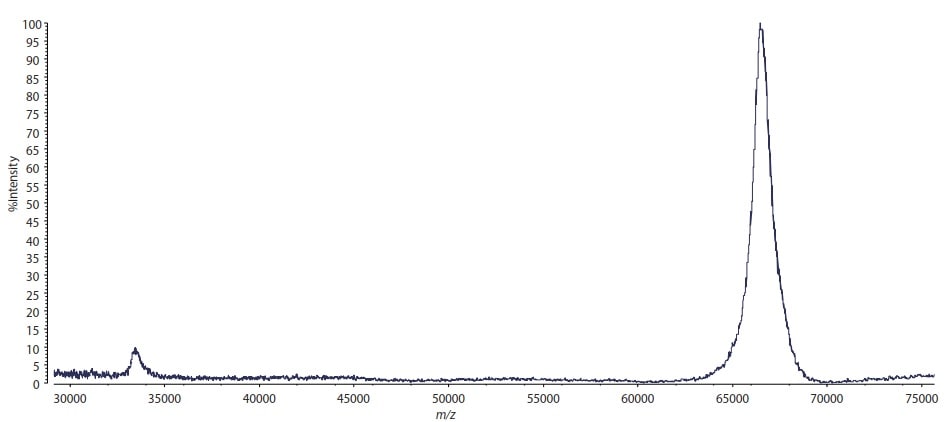
250 fmol (approx. 17 ng) bovine serum albumin (BSA) was mixed with sinapinic acid (10 mg/mL, 50 % acetonitrile/0.1 % trifluoroacetic acid solution) and analyzed in linear mode using MALDI-8020. The result is shown in Fig. 1. The peak of the BSA's singly-charged protonated molecule is detected at m/z 66430 with S/N > 100.
Benchtop MALDI-TOF MS "MALDI-8020"

Key features:
- Linear mode (positive ion) MALDI-TOF
- 200 Hz solid-state laser, 355 nm
- Load-lock chamber for fast sample introduction
- UV laser-based source cleaning (patented)
- Small footprint/benchtop design
- Quiet operation (<55 dB)