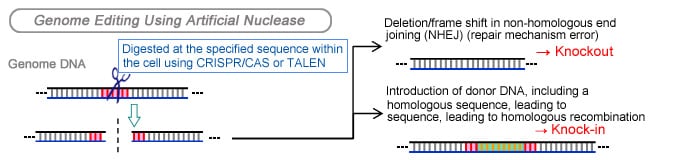
Gene Knockout, Genome Editing
Genetic Analysis of Knockout Animals
Knockout animals are defined as animals in which the sequence coding a target gene has been artificially deleted. They are analyzed in order to compare their genetic function to the normal population, and to create model animals with diseases caused by missing genes.
Confirming Genetic Knockout Using Genome Editing Tool

The emergence of genome editing tools such as CRISPR/CAS and TALEN has allowed the deletion and insertion of sequences using a method that is much simpler than those conventionally used. Post-editing confirmation by DNA sequencing, however, has been problematic costing both time and money.
The following is an example of an extremely simple procedure using the MCE-202 MultiNA microchip electrophoresis system for DNA/RNA analysis, which confirms the presence or otherwise of deletion or insertion occurring in the target gene.
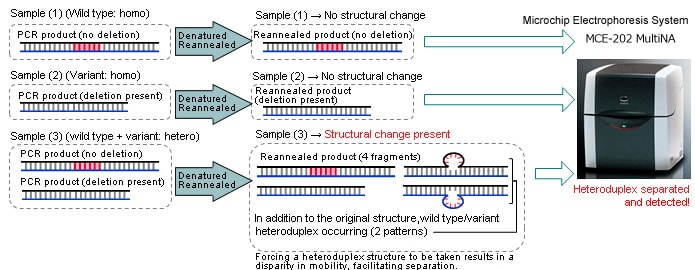
Analysis Principles/Method
Subsequent to genome editing, the region including the deleted point is amplified by PCR. As shown below, the PCR products are denatured and then reannealed to form a heteroduplex. The structural change results in a different migration pattern, which is measured by the MultiNA to confirm the presence of the deletion. This method has the additional advantage of allowing the identification of short deletions, which are difficult to determine Fragment length difference.
Analysis Example
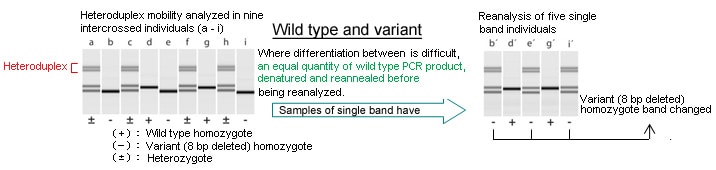
Identification of homo/heterozygous variant introduced via TALEN
Medaka Oryzias laptipes (heterovariant) was intercrossed -with TALEN-introduced variant (with 8 bp deleted), and PCR was performed on the deletion region of the population produced, after which heteroduplex mobility was analyzed using the MultiNA.

Analysis method/data supplied by: Masato Kinoshita, PhD, Graduate School of Agriculture, Kyoto University
Citation: Ansai, S. et al., Biol Open. 2014 Apr 11;3(5):362-71.(CRISPR)
Citation: Ansai, S. et al., Dev Growth Differ. 2014 Jan;56(1):98-107.(TALEN)
Rat Genotyping from Blood Using Ampdirect
Ampdirect counteracts PCR inhibitors such as protein and sugars, facilitating direct PCR from samples including from a crude state, without the need to refine template DNA. Below, we introduce an example of swift genotyping from rat blood applied to an FTA® card, using Ampdirect and the MultiNA.
Analysis Example
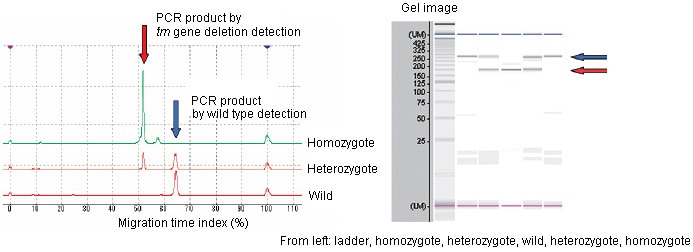
SERs (spontaneously epileptic rats) are model animals for human epilepsy which causes severe absence seizures and tonic convulsions. The SERs were developed at the Institute of Laboratory Animals, Graduate School of Medicine, Kyoto University by hybridizing the genes in two mutant strains of tremor rat and zitter rat, which are homozygous for both tremor (tm) and zitter (zi) mutations. The tm mutation is a genomic deletion (roughly 200 kbp) in theAspa(aspartoacylase) gene, while zi is an 8-bp deletion in theAtrn(attractin) gene. The strain is maintained by sibling mating of a zitter homozygote with a tremor heterozygote. These deficiencies must be detected in order to select the parent rats for creating the next generation. Of the two mutant genes, the tm mutation presents a long deletion of approximately 200 kbp, and multiplex PCR using two pairs of primers is conducted to detect the presence or absence of that deficiency. One pair is for wild-type detection, in which amplification is done using primers that are annealed in this deletion region, resulting in PCR amplification product only if no deletion exists. The other pair for tm gene deletion detection is annealed across the region containing deletion, with the result that a PCR amplification product (approx. 240 bp) is obtained only if a deletion exists. If no deletion exists, no PCR amplification product is obtained, since the region is extremely long (approx. 200 kbp).
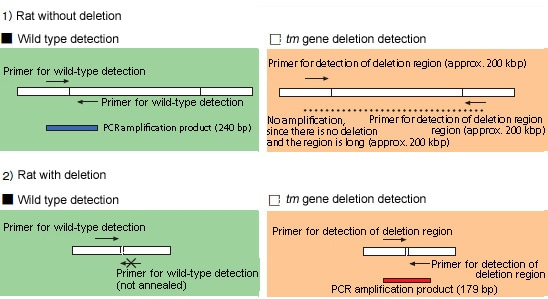
This can be summarized as in the table below.
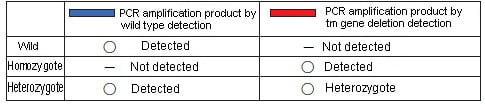
Results
Analysis by MultiNA (Kit used: DNA-500 Pre-Mix Mode)
|
Samples supplied by: |
Associate Professor Takashi Kuramoto, Institute of Laboratory Animals, Graduate School of Medicine, Kyoto University |
Microchip Electrophoresis System for DNA/RNA

The Microchip Electrophoresis System is effective for quick and easy checking of the size of DNA or RNA nucleic acid samples and for approximate quantitation. Reusable high-performance microchip achieves lower analysis costs than agarose gel electrophoresis. Simply load the samples and separation buffer to perform up to 120 unmanned, automated analyses.


