Quality Control for next-generation DNA sequencing
Due to technological advancements in next-generation sequencers, they are being used for a wider range of applications, such as for de novo sequencing or for the analysis of mutations, exomes, and expressions. Furthermore, it is possible to select the model based on the throughput required for data analysis and data processing, which has increased their use rapidly. However, to obtain good sequencing results, regardless of the application or model, it is necessary to validate the size distributions and concentrations in the NGS library. Therefore, quality control (QC) of that information is essential when using NGS systems.
The processes required, from library preparation to QC, involved a series of tedious manual operations. This article describes an example of mouse RNA sequencing analysis that solves this problem by using an MCE-202 MultiNA automatic electrophoresis system for NGS library QC.
Sequencing Process Flow and Range of MultiNA Applications
During the NGS library QC process, MultiNA's smear analysis software* can be used to calculate the estimated average library size, concentrations, and mole concentrations.
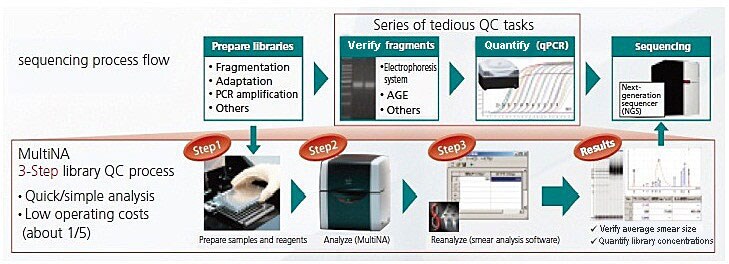
NGS Process Flow and Range of MultiNA Applications in Library QC
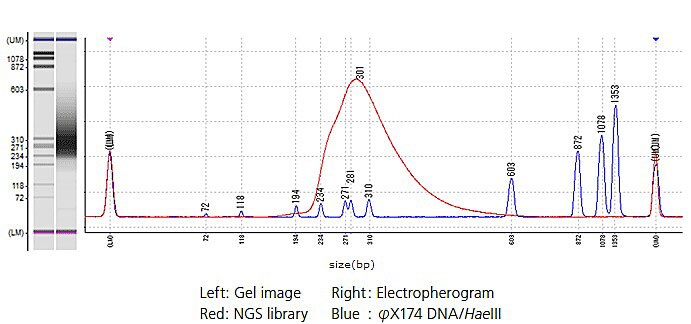
Example of NGS Library Electrophoresis
The mouse RNA sequencing
The mouse RNA sequencing results showed adequate read length and a stable read ratio between respective libraries due to index tag sequencing. These results indicate that library QC using the MultiNA can be used not only to verify size distributions, but also to simultaneously quantify libraries under routine analysis. By using the MultiNA system for library QC, it is possible to perform QC quickly and easily.
| Cluster Density | 548 K / mm2 | Total Read Count | 151.5 M reads |
| Read Count (after filtration) |
143.3 M reads | ≥ Q30 ratio | 95.9 % |
| Read Ratio of 4 Libraries | 21.2 - 26.2 % (Four libraries per lane sorted by index tag sequencing: Theoretical value of 25 %) |
||
* MultiNA software requires version 1.12 or later.
Microchip Electrophoresis System for DNA/RNA

DNA and RNA samples are separated by size by the electrophoresis system using a microchip so that the size of nucleic acid (DNA/RNA) samples is verified and approximately quantitated. The microchip achieves an electrophoresis system capable of speedily conducting electrophoresis separation, and a fluorescent detector ensures that analysis is performed to high sensitivity and, moreover, fully automatically.


